Central Dogma
George Beadle and Edward Tatum in 1941 studied Neurospora crassa [Fungi], which is a type of red bread mold of the phylum Ascomycota.
The spores of the fungus Neurospora crassa were exposed to mutagenic agents like X-rays causing mutations in DNA.
They obtained some mutant strains of Neurospora crassa which were lacking in specific enzymes.
They concluded that a gene is a segment of DNA which codes for an enzyme.
It was named as One gene-One enzyme hypothesis.
Later in 1957 this hypothesis was modified by Vernon Ingram as One-gene-One polypeptide hypothesis.
It is because of the fact that many genes code for proteins which are not enzymes.
Protein synthesis is the most important, essential and significant metabolic activity of the living world taking place in every living cell continuously.
For actual phenotypic expression of any character in a living cell or in the body of an organism biochemical reactions are essential.
Each biochemical reaction needs a specific enzyme for its initiation and completion.
Almost all the enzymes are proteins.
The cell also needs many other structural proteins.
Thousands of different types of structural and catalytic proteins are continuously required within the cell at any moment.
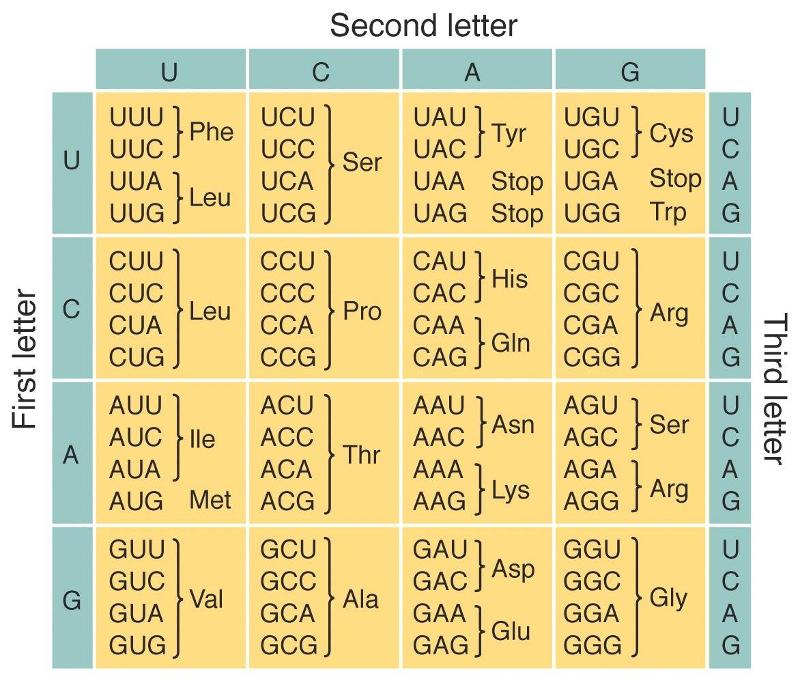
The Genetic Code
DNA contains all the genetic information of an organism.
The genes control, regulate and express the characters.
The expression of a character takes place through- a specific protein.
The gene is a segment of DNA, mRNA molecule is formed on DNA which later helps in construction of a polypeptide chain.
Thus, the mRNA acts as an agent in conveying information from DNA to cytoplasm.
A sequence of three nucleotides on the mRNA strand is called codon.
Each codon codes for a specific amino acid.
This was suggested by George Gamow in 1954 while the direct evidence for this was provided by Crick in 1961.
Subsequently Marshall Nirenberg, Heinrich Matthaei and Har Gobind Khorana deciphered (cracked) complete genetic code.
They used artificial mRNA templates, (homopolymers and copolymers), and cell free system of protein synthesis.
For this work Khorana shared Nobel Prize in 1968 with Nirenberg and Holley.
In RNA. there are four types of nitrogen bases (A.U,G,C).
They can form 64 possible combinations of triplets, called codons.
Among these 3 codons serve as termination codons and remaining 61 codons are sense codons.
They code for 20 essential amino acids required for protein synthesis.
Characteristics of genetic code:
1. Genetic code is triplet and commaless i.e. on the mRNA strand the triplet codons are arranged one after the other without any gap or space.
2. It is non-ambiguous. Each codon will specify a particular amino acid.
Exception.
- AUG codes for methionine
- GUG codes for valine
- if AUG is not available then GUG codes for methionine, as a start codon in protein synthesis.
3. The genetic code is degenerate, as 61 codons are available for 20 amino acids.
- Two or more codons can specify the same amino acid. Hence it is said to be degenerate e.g. GGG GGA. GGC and GGU code for glycine.
- Polarity is one of the important feature of the genetic code, it can be read only in 5' to 3' direction of m-RNA.
- The genetic code has start and stop signals.
The codon AUG acts as start / initiation codon which codes for methionine.
The codons UAG(amber). UAA (ochre) and UGA (opal) serve as stop codons or termination codons.
- The genetic code is universal i.e. it is similar in all the organisms, from simple bacteria to complex organisms.
However, some exceptions are found in mitochondria of Yeast and Mycoplasma.
Degeneracy of Code Explained by Crick in 1966
There are multiple codons for one amino acid.
There are 61 codons for 20 amino acids.
One would expect presence of 61 types of tRNAs, each with a specific anticodon for recognition of a particular codon.
However, the actual types of tRNAs is much less than 61.
This indicates that the anticodon of tRNA can read two or more codons on mRNA.
In 1966 Crick proposed Wobble hypothesis to explain the same.
According to this hypothesis, in codon-anticodon pairing the third base may not be complementary.
The third base of the codon is called wobble base and this position is called wobble position.
The actual base pairing occurs at first two positions only.
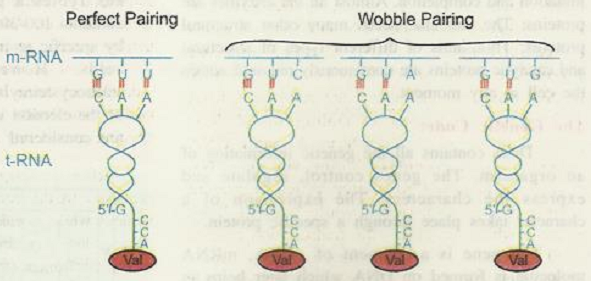
In the above example though the codon and anticodon do not match perfectly then also the required amino acid is brought perfectly.
This enables the economy of tRNA. GUU. GUC. GUA and GUG code for amino acid - Valine.
Why each codon present in the genetic code should have 3 bases only and not one or two bases?
We know that there are in all 20 different types of essential amino acids required for synthesis of proteins in the cells.
Each amino acid will require al least one specific codon. Thus there should be al least 20 codons in the genetic code which is composed of only four bases A. U. G and C.
If each codon has only one base/nucleotide then there will be 41 = 4 codons are possible (not enough to code 20 different amino acids).
If each codon has two bases/nucleotides-then there will be 42 = 4x4 = 16 codons are possible (not enough to code 20 different amino acids)
If each codon has three bases/nucleotides -then there will be 43 = 4 x 4 x 4 = 64 codons are possible (more than enough to code 20 different amino acids)
Central dogma of protein synthesis
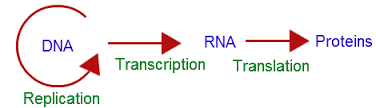
In 1958 Crick proposed that DNA determines the sequence of amino acids in a polypeptide (protein) through mRNA.
This is the main principle or central dogma of protein synthesis which involves transcription and translation.
This process needs;
i) DNA to provide the base sequence,
ii) 20 types of amino acids.
iii) Non-genetic types of RNA (mRNA, rRNA. tRNA).
iv) Ribosomes as site of protein synthesis,
v) Various enzymes/factors and
vi) ATP/GTP as source of energy
Transcription
It is formation of mRNA on DNA template.
The process takes place in presence of DNA dependent RNA polymerase.
In this process genetic information from DNA is copied into RNA. (Transcribe means copy in writing).
Therefore the process is called transcription.
The script i.e. genetic information is now with mRNA.
For the transcription activity, a transcription unit works which has
i) Promoter,
ii) The structural gene and
iii) A Terminator.
The DNA strand which is used for synthesis of RNA is called antisense or template strand which is oriented in 3' to 5' direction.
The Other strand, not involved in RNA synthesis is called sense or coding strand. It is oriented in 5' to 3' direction.
Promoter is a small DNA sequence which provides binding site for RNA polymerase, which is present towards 5' end / upstream.
Terminator is small DNA sequence which terminates the transcription process, which is present towards 3 end /downstream.
(Here the polarity of coding strand is considered as reference).
The structural gene which is to be transcribed is
- monocistronic in eukaryotes
- polycistronic in prokaryotes.
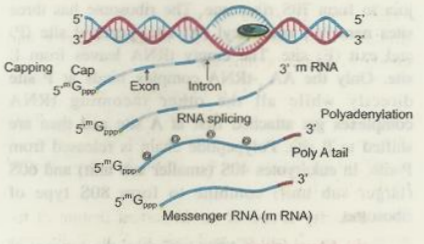
In eukaryotes the genes are split, having exons and introns.
The DNA sequences which are expressed or appear in the mature or processed RNA are the exons while those, not appearing in mature or processed RNA are the introns.
During transcription the enzyme RNA polymerase binds to the promoter site and brings about the initiation of the process.
The two strands of DNA separate from each other.
According to the base sequence present on the template strand, the complementary RNA nucleotides are selected and joined one after the other to form the mRNA strand (elongation).
A small part of RNA remains attached to the enzyme. As the enzyme reaches to the terminator region, both the enzyme and newly constructed RNA fall off.
This is the termination of transcription. Initiation factor (sigma) and termination factor (rho) play an important role in the initiation and termination.
In the prokaryotic organisms as bacteria, the newly formed RNA do not require further processing, but in eukaryotes the matter is different.
In eukaryotes. there are three types of RNA polymerases;
- RNA polymerase - I for formation of rRNA.
- RNA polymerase - II for synthesis of precursor of mRNA i.e. heterogeneous nuclear RNA (hnRNA) and
- RNA polymerase - III for formation of tRNA. snRNA (small nuclear RNA).
In eukaryotes. the RNA is non-functional when it is formed and undergoes splicing, capping and tailing.
The removal of introns from the RNA is called splicing.
Addition of an unusual nucleotide methyl guanosine triphosphate at 5"end of hnRNA (having both the introns and exons) is called capping.
Addition of adenylate residues at 3' end is called tailing.
The hnRNA which has undergone capping. splicing and tailing now functions as mRNA.
After transcription
In prokaryotes, the mRNAs are used in the same compartment as there is no nucleus.
In eukaryotes they are produced in the nucleus and then are transported out of the nucleus to the ribosomes (in cytoplasm) which is a site for protein synthesis i.e. translation.
Translation
Translation: It is the process in which the sequence of codons on the mRNA strand is used (read/decoded) and accordingly the amino acids are joined to each other to form a polypeptide chain that makes protein.
The process involves following su-steps;
(A) Activation of amino acids and formation of AA-tRNA complex:
In presence of an enzyme amino acyl tRNA synthetase the amino acid (AA) molecule is activated and then each amino acid is attached to the specific tRNA molecule at 3' CCA end to form aminoacyl-tRNA complex.
The reaction needs ATP. This process is called charging of tRNA or aminoacylation of tRNA.
(B) Formation of the polypeptide chain
It is an actual translation which involves following steps ;
(i) Initiation:
It begins with the formation of initiation complex which requires the mRNA having codons for a polypeptide, the smaller (30S) and larger (50S) sub-units of ribosome.
the initial AA, -tRNA complex and ATP and GTP as source of energy.
The process of initiation needs transcription factors.
In prokaryotes the first AA - tRNA complex has amino acid, N-formyl- methionine (f-met) while in eukaryotes It is methionine (met).
The process starts with binding of mRNA on the smaller 30S sub-unit of ribosome.
The start codon AUG is positioned properly. The AA tRNA complex i.e. f-met-tRNA complex now gets attached to the start codon AUG.
This is done with the help of anticodon UAC of tRNA.
Small and large unit of ribosome join lo form 70S ribosome.
The ribosome has three sites namely :
1. amino acyl (A) site - incoming AA-tRNA complexes get attached
2. peptidyl site (P) - the AA -tRNA complex binds
3. exit (E) site - empty tRNA leaves from E site
Polypeptide chain is released from P-site.
In eukaryotes 40S (smaller sub unit) and 60S (larger sub unit) combine to form 80S type of ribosome.
(ii) Elongation
After initiation, the process of elongation starts.
This is done by formation of peptide linkages/bonds in between the successive amino acid molecules (AAI, AA2. AA3 and so on ->).
The elongation activity is catalyzed by the enzyme peptidyl transferase.
Each tRNA complex brings a specific amino acid.
Due to complementary nature of anticodons and codons the amino acids are placed to their proper positions.
The elongation activity involves elongation factors.
During elongation, the ribosome moves along the mRNA in step wise manner from start to stop codon (5' -> 3') one codon ahead each time.
This movement is called translocation. In every step of translocation one amino acid is added in the polypeptide chain causing elongation.
(iii) Termination;
Elongation continues until the ribosome adds the last amino acid coded by the mRNA.
When the mRNA reaches to the last termination codon. i.e. either UAA. UAG or UGA, termination occurs.
In identifying the stop/termination codon and in releasing the polypeptide chain from the site, the release or termination factors R1, R2 and S play an important role.
After termination, the smaller (30S) and larger (505) sub units of ribosome get separated from each other.
The energy requirement for (polypeptide chain) protein synthesis activity is fulfilled by ATP and GTP.
Polysomes or Polyribosomes -
In order to increase the cellular efficiency of protein synthesis, many ribosomes may bind to the mRNA strand and form the polypeptide chains for synthesis of protein molecule.
Such a structure with many ribosomes bound to mRNA is called polysome or polyibosome.
Each ribosome brings about synthesis of polypeptide chain and many identical chains are produced.
Ribosomes are composed of approximately 65% r-RNA and 35% Protein.
They are not surrounded by any membrane. The two unequal and irregularly shaped sub-units fit together.
It form a cleft during protein synthesis through which the m-RNA passes as ribosome moves along it during translation.
