Restriction Digestion:
RE Digestion : Theory
Restriction digestion is the enzymatic techniqueused for the cleavage of DNA molecules at specific sites so that all the DNA fragments containing a particular sequence have the same size. This cleavage method is performed by the use of DNA cleaving enzymes of bacteria. This group of enzymes called restriction enzymes are capable of cleaving DNA molecules at particular short-base sequences located at a particular position on the DNA. This technique is sometimes also referred to as DNA fragmentation.
Nowadays each molecular biology experiment is coupled with restriction digestion for wider applications in the field of cloning as well as other analytical techniques.
The most common use of restriction digestion is the cloning of a DNA fragment into a suitable vector such as a cloning and/or expression vector. Other important applications of restriction digestion include restriction mapping, analysing population dynamics, rearranging DNA molecules, preparation of molecular probes, creating mutants and many others.
Restriction enzymes are known to make double-strand cuts in DNA molecules.
Under natural conditions, bacteria secrete these enzymes to defend themselves against the foreign invading DNA from other bacteria and viruses.
Now restriction enzymes have become an important part of the molecular biology toolbox for wide applications.
Restriction enzymes fall under the group of nucleases that cleave the sugar-phosphate back bone of a DNA molecule.
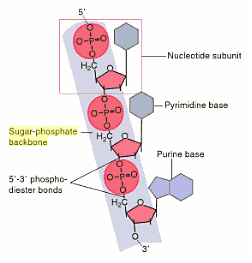
The restriction enzymes which cut within the DNA molecule are called restriction endonucleases.
They can cleave the DNA molecules at specific points called restriction sites to generate a set of smaller fragments.
There are three types of restriction enzymes based upon the restriction and modification systems.
• Type I Enzymes:
They exhibit both restriction digestion and modification which require cofactors for their activity.
1. Mg2+ ions,
2. S-adenosylmethionine (SAM) and
3. adenosine triphosphate (ATP).
Type I restriction enzymes cleave DNA at non-specific sites which may be 1000 bp. or more.
As methylation reaction is also performed by the same enzyme and the target DNA is modified prior to cutting, this type of restriction enzyme is of less value in gene manipulation practices.
• Type II Enzymes:
Type II enzymes possess two separate proteins for restriction cleavage and modification.
In this case, the restriction activity is not dependent on cofactors such as ATP or SAM.
The only requirement as cofactors in this case is Mg2+ ions.
The major advantage of this group of enzymes is their site specific nature to hydrolyse specific phosphodiester bonds in both DNA strands. Hence, this group of restriction enzymes is used for the purpose of restriction digestion and recombinant technology.
In most cases it is also used in
1. genome mapping,
2. restriction fragment length polymorphism (RFLP),
3. sequencing and
4. cloning.
• Type III Enzymes
These enzymes are similar to that of type I systems, possessing both restriction digestion and modification activities simultaneously.
They can recognise and cleave 25-27 bp outside the recognition sequence downstream to 3' direction and also require Mg2+ for their activity.
Type II Enzymes - Cleavage Patterns
Type II restriction enzymes generate three types of DNA ends possessing 3' → 5' phosphodiester bond and 3'-hydroxyl groups.
1. cohesive 5' ends. e.g. EcoRI
2. cohesive 3'ends. e.g. PstI
3. blunt ends. e.g. Haelll.
Plasmids act as useful vectors for transferring the genetic material from one organism to another by recombinant DNA technology.
Restriction endonuclease is used in this process to cut and insert foreign DNA pieces into plasmid vectors.
There are many factors that affect the activity of restriction enzymes in vitro which include
1. temperature,
2. buffer systems,
3. ionic conditions
4. methylation of DNA.
Most digestions are generally carried out at 37°C.
Certain restriction enzymes like SmaI require lower temperatures (25°C),
Some restriction enzymes require higher temperatures (TaqI 65°C).
A buffer system is mandatory for the restriction action of the enzymes as most of them have the optimum activity at a range of pH 7.0-8.0.
Most restriction enzymes also require Mg2+ for their activity; however, in some cases either ions like Na+ and K+ are also required, which depends on the nature of the enzyme.
Methylation of specific adenine or cytidine residues affects the action of the restriction enzymes in an adverse manner.
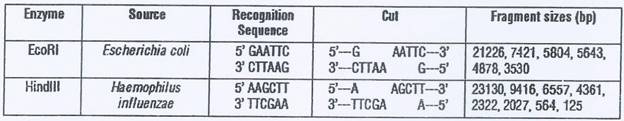
Every restriction enzyme has unique target sites for digestion.
Lambda DNA has multiple restriction sites for both EcoRI and HindIII which result into several fragments of varying sizes.
Restriction Digestion: Experiment
Requirements.
Lambda DNA, DNA marker, agarose, EcoRI, Hindlll, 10X assay buffer for EcoRI, 10X assay buffer for HindIII, molecular biology grade water, 50X TAE, 6X gel loading buffer, polypropylene tubes (0.5 ml), measuring cylinder, beaker, ethidium bromide (10 mg/ml), electrophoresis apparatus, UV trans-illuminator, heating block or water bath, vortex mixer, micropipettes, tips, adhesive tape, crushed ice, microwave/ hotplate/ burner
Protocol
1. Setting up the RE reactions:
a. Before starting the experiment, crush ice and place the vials containing Lambda DNA, Restriction Enzymes and Assay Buffers onto it.
b. In this experiment Lambda DNA is digested with two restriction enzymes; EcoRI and HindIII.
c. Set up the reaction mixture as follows :
Reaction 1 (EcoRI digestion) :
1. Lambda (λ) DNA - 5.0 μl
2. 10X Assay Buffer of EcoRI - 2.5 μl
3. Milli Q water - 16.5 μl
4. EcoRI - 1.0 μl
Reaction 2 (HindIII digestion):
1. Lambda (λ) DNA - 5.0 μl
2. 10X Assay Buffer of HindIII - 2.5 μl
3. Milli Q water - 16.5 μl
4. HindIII - 1.0 μl
Total - 25 μl
d. After preparing the two reaction tubes, mix the components by gentle pipetting and tapping.
e. Incubate the tubes at 37°C for 1 hour.
f. After 1 hour incubation, immediately place the vials at room temperature (15-25°C) for 10 minutes.
g. After incubation run the samples on agarose gel.
2. Agarose Gel Electrophoresis :
a. Preparation of 1X TAE :
To prepare 500 ml of 1X TAE buffer add 10 ml of 50X TAE Buffer to 490 ml sterile distilled water (MilliQ water recommended).
b. Preparation of agarose gel :
To prepare 50 ml of 1% agarose gel, measure 0.5 g agarose in a glass beaker or flask and add 50ml 1X TAE buffer.
- Heat the mixture on a microwave or hot plate or burner, swirling the glass beaker/flask occasionally, until agarose dissolves completely (Ensure that the lid of the flask is loose to avoid buildup of pressure).
c. Loading of the DNA samples :
Load 10 μl of ready to use DNA Marker into the well 1.
To prepare samples for electrophoresis, add 2 μl of 6X gel loading buffer to 10 μl of DNA samples. Mix well by pipetting and load the samples into the well.
d. Electrophoresis :
Connect the power cord to the electrophoretic power supply according to the conventions: Red-Anode and Black-Cathode.
Electrophorese at 100-120 V and 90 mA until dye markers have migrated an appropriate distance, depending on the size of DNA to be visualized.
3. Interpretation of Gel
Following electrophoresis of the digested samples on agarose gel, look for the digestion pattern for the two restriction enzymes.
Compare the size of each fragment,with that of the DNA marker.
Restriction digestion patterns of lambda DNA obtained upon treatment with EcoRI and HindIII are markedly different which demonstrates the fact that each restriction enzyme recognizes and cleaves only a specific base sequence unique to it.
The size of the fragments can be determined by comparing with that of the DNA marker ran on the same gel.
Result

