Ligation
When two sets of DNA fragments are mixed, base pairing between sticky ends results in the coming together of fragments from different or same molecule.
Such pairings are transient owing to the weakness of H-bonding between the few bases in sticky ends.
Construction of recombinant DNA molecule is therefore dependent on the ability to covalently seal single stranded nicks in DNA.
The process of sealing single stranded nicks in DNA is accomplished both in vivo and in vitro by the enzyme DNA ligase. It catalyses the formation of phosphodiester bonds between juxtaposed 5' phosphate and a 3' hydroxyl terminus of double stranded DNA. It can repair single stranded nicks in dsDNA and joins dsDNA restriction fragments having either blunt ends or homologous cohesive ends.
One cohesive end ligation unit is defined as the amount of enzyme required to give approximately 50% ligation of 1μg of lamda DNA / HindIII digest in 30 minutes at 16°C in a 20 μl reaction mixture at a 5' termini concentration of 0.12 μM (300 μg/ml).
1 cohesive end ligation unit approximately equals 0.015 ATP-PP exchange units (Weiss unit).
One Weiss unit is equivalent to -200 cohesive-end ligation units.
E. coli ligase and T4 DNA ligase are the two DNA ligases used in nucleic acid research. They differ in their requirement for energy source and in their ability to ligate blunt ends.
The enzyme isolated from E. coli is a polypeptide with a molecular weight of 75kD. It requires NAD+ as the cofactor.
T4 DNA ligase is approximately 68000 Dalton (68kD) protein produced by bacteriophage T4 (gene 30) requiring ATP as energy source.
E. coli ligase is inhibited by trypsin while T4 ligase is inhibited by high salt concentration (NaCl or KCl if the concentration >200 mM).
T4 ligase is obtained from an induced lysogen of T4 lig phage. It has the unique ability to join sticky and blunt ended fragments.
Cohesive end ligation is carried out at 12°C to 16°C to maintain a good balance between annealing of ends and activity of the enzyme and PEG is added to the reaction mixture for stimulation.
If reaction is set at higher temperatures annealing of the ends become difficult, while lower temperatures diminishes the ligase activity.
An advantage with T4 enzyme is that it can quickly join and produce a full complement, but it is difficult to retrieve the inserted DNA from vector.
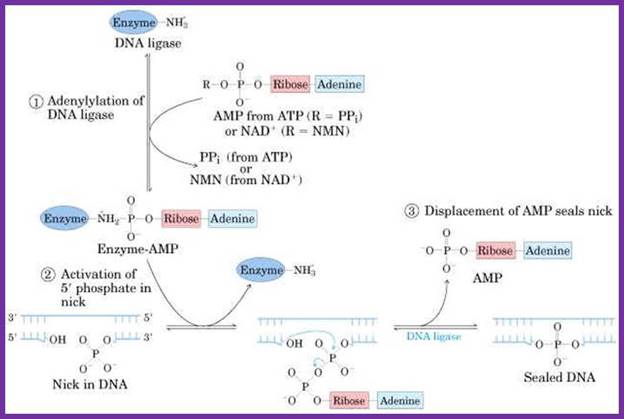
E. coli as well as T4 DNA ligase contain an amino group on lysine residue. In both enzymes, the cofactor breaks into AMP which in turn adenylates the enzyme to form an enzyme-AMP complex (EAC).
EAC binds to nicks containing 3'-OH and 5'-PO4 ends on a dsDNA molecule.
The 5'-PO4 terminus of the nick is adenylated by the EAC with 3'-OH terminus resulting in the formation of a phosphodiester bond with the liberation of AMP resulting in the sealing of the nick.
The ligation reaction is often carried out at 4°C to lower the kinetic energy of the molecule. This reduces separation of paired sticky ends which are stabilized by ligation. However, very long reaction times are needed to compensate for the low activity of DNA ligase in the cold.
The lack of cohesive termini makes blunt end ligation more complex and significantly slower. Since annealing of ends is not a factor, the reaction is carried out at 24°C. 10-100 times more enzyme is required to achieve similar ligation efficiency as that of cohesive end ligation. However, blunt end ligation is a useful way of joining together DNA fragments which have not been produced by the same RE; which therefore probably have incompatible sticky ends. These ends are first removed prior to ligation by using S1 nuclease which can digest ssDNA.
Requirements for ligation :
Lambda DNA - Hindlll digest, 10X ligase assay buffer, T4 DNA ligase, molecular biology grade water, agarose, 50X TAE buffer, 6X gel loading buffer, 0.5 ml polypropylene tubes, measuring cylinder, beaker, ethidium bromide (10 mg/ml), electrophoresis apparatus, UV transilluminator, water bath, micropipettes, tips, adhesive tape, crushed ice, microwave/ hotplate/ burner.
Ligation : Experiment
1. Setting up the ligation reaction:
a. Before starting the experiment, crush ice and place the vials containing Lambda DNA-Hindlll digest, 10X ligase buffer andT4 DNA ligase onto it.
b. In this experiment Lambda DNA-Hindlll digest is ligated with T4 DNA ligase.
c. Set up the reaction mixture as follows:
1. Lambda (A) DNA-Hindlll digests - 4.0 μl
2. 10X Ligase Assay Buffer - 1.0 μl
3. Milli Q water - 4.0 μl
4. T4 DNA Ligase - 1.0 μl
Total - 10 μl
d. After preparing the reaction tube, mix the components by gentle pipetting and tapping.
e. Incubate the tubes at 16°C waterbath for 3 hours.
f. After incubation run the samples on agarose gel.
Result

