4.3 DNA packaging :
Length of DNA double helix molecule, in a typical mammalian cell is approximately 2.2 meters.
This can be worked out simply by multiplying the total number of base pairs with distance between the consecutive base pairs.
Approximate size of a typical nucleus is 10-6m.
Q. How this long DNA molecule can be then accommodated in such a small nucleus?
Ans: It, therefore, must be condensed, coiled and super-coiled to fit inside such small nucleus.
Packaging in Prokaryotes:
In prokaryotes like E. coli, cell size is almost 2-3μm long.
They do not have well organized nucleus. It is without nuclear membrane and nucleolus.
- The nucleoid is small, circular, highly folded, naked ring of DNA which is 1100μm long in perimeter, containing about 4.6 million base pairs.
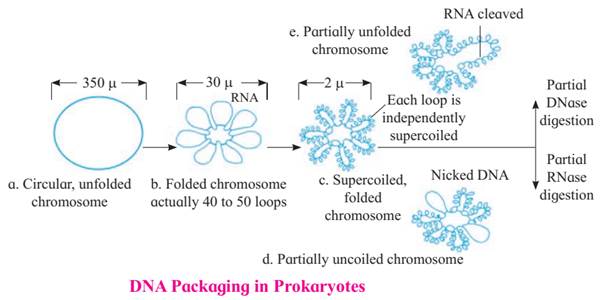
o The nucleoid, a 1100μm long DNA molecule, needs to fit into a 2-3μm cell.
o To reduce size, the DNA circularizes, decreasing its diameter to 350μm.
o Further compaction occurs through folding and looping, reducing the diameter to 30μm.
o 40-50 loops or domains are formed with the help of RNA connectors.
o Each loop undergoes coiling and supercoiling, reducing the diameter to 2μm.
o Proteins like HU and enzymes like DNA gyrase and topoisomerase I assist in supercoiling and maintaining the compact
Packaging in Eukaryotes:
Eukaryotes show well organized nucleus containing nuclear membrane, nucleolus and thread-like material in the form of chromosomes.
In the chromosomes, DNA is associated with histone and non-histone proteins as was reported by R. Kornberg in 1974.
The organization of DNA is much more complex in eukaryotes. Depending upon the abundance of amino acid residues with charged side chains, a protein acquires its charge.
Histones are the proteins that are rich in lysine and arginine residues. Both these amino acid residues are basic amino acids and carry positive charges with them. So, histones are a set of positively charged, basic proteins (histones + protamine).
These histones organize themselves to make a unit of 8 molecules known as histone octamer.
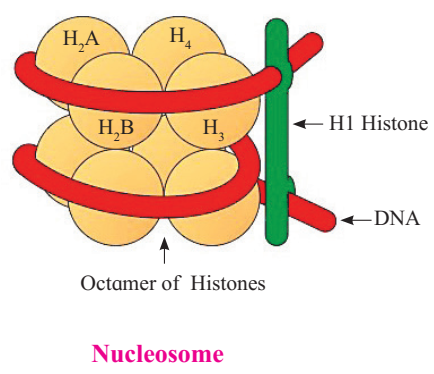
The negatively charged helical DNA is wrapped around the positively charged histone octamer, forming a structure known as nucleosome. The nucleosome core is made up of two molecules of each of four types of histone proteins viz. H2A, H2B, H3and H4. H1 protein binds the DNA thread where it enters (arrives) and leaves the nucleosome.
One nucleosome approximately contains 200 base pair long DNA helix wound around it. About 146 base pair long segment of DNA remains present in each nucleosome.
Nucleosomes are the repeating units of chromatin, which are thread-like, stained (coloured) bodies present in nucleus. These look like 'beads-on-string', when observed under an electron microscope. DNA helix of 200 bps wraps around the histone octamer by 1¾ turns.
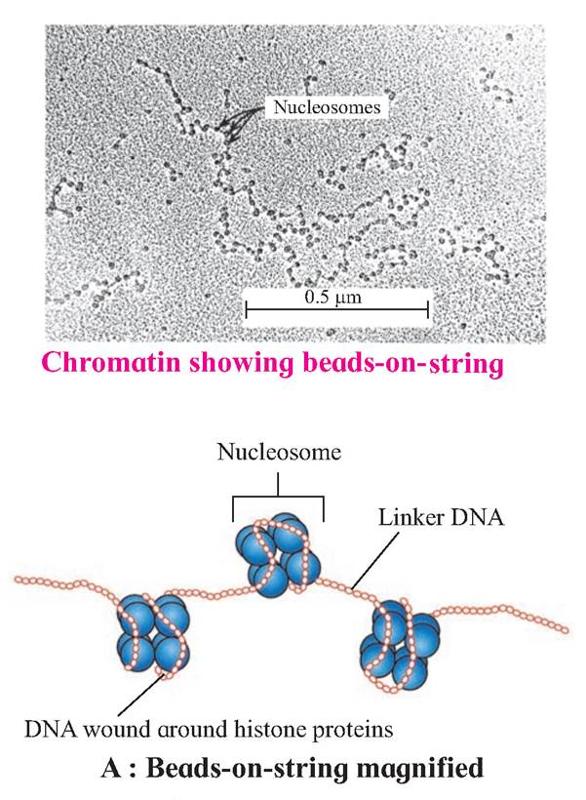
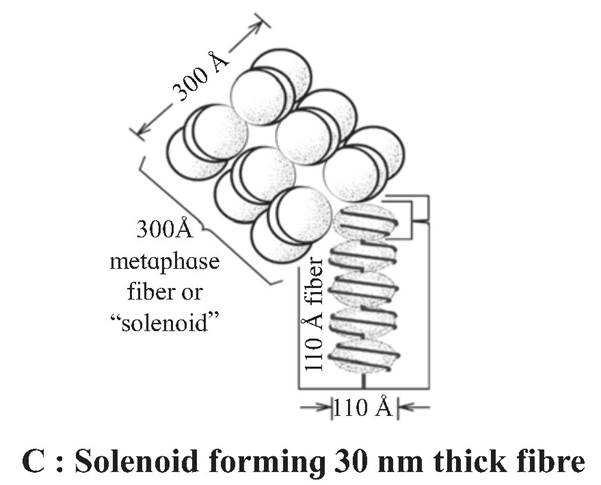
Six such nucleosomes get coiled and then form solenoid that looks like coiled telephone wire.
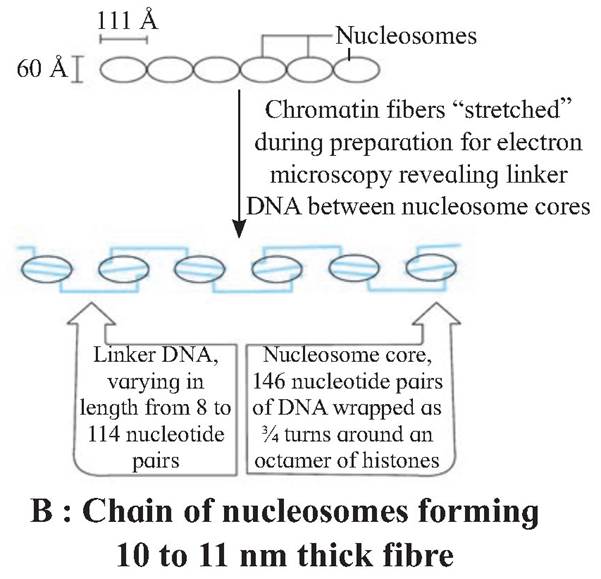
The chromatin is packed to form a solenoid structure of 30 nm diameter (300A) and further supercoiling tends to form a looped structure called chromatin fiber, which further coils and condense at metaphase stage to form the chromosomes.
The packaging of chromatin at higher levels, need additional set of proteins that are called Non-Histone Chromosomal proteins (NHC).
Non-Histone Chromosomal Proteins (NHC) :
These are additional sets of proteins that contribute to the packaging of chromatin at a higher level.
Heterochromatin and Euchromatin :
1. Heterochromatin:
In eukaryotic cells, some segments of chromonema/ chromosome during interphase and early prophase remain in a condensed state. These region constitute heterochromatin. This term was proposed by Heitz. These regions are localized near centromere, telomeres and are also intercalated. It is genetically mostly inactive. It stains strongly and appears dark. Heterochromatin is 2 to 3 times more rich in DNA than in the euchromatin.
2. Euchromatin:
The regions of chromonema which are in non-condensed state, constitute euchromatin. Euchromatic regions stain light. Euchromatin is genetically very much active and fast replicating. Euchromatin is transcriptionally active, while heterochromatin is transcriptionally almost inactive.
