4.4 DNA Replication :
The DNA molecule regulates and controls all the activities of the cell. Because of its unique structure, it is able to control the synthesis of other molecules of the cell. At the same time when the cell reproduces, the DNA also should duplicate itself to distribute equally to the daughter cells. As a carrier of genetic information, DNA has to perform two important functions :
a. Heterocatalytic function :
When DNA directs the synthesis of chemical molecules other than itself, then such functions of DNA are called heterocatalytic functions.
Eg. Synthesis of RNA (transcription), synthesis of protein (Translation), etc.
b. Autocatalytic function :
When DNA directs the synthesis of DNA itself, then such function of DNA is called autocatalytic function.
Eg. Replication. The process by which DNA duplicates itself is called replication. Through replication, it forms two copies that are identical to it.
Semi-conservative Replication of DNA
In eukaryotic organisms, replication of DNA takes place only once in the cell cycle. It occurs in the S- phase of interphase in the cell cycle.
DNA replicates through Semiconservative mode of replication. The model for Semiconservative replication was proposed by Watson and Crick, on the basis of antiparallel and complementary nature of DNA strands.
The process of semi-conservative replication is as below:
1. Activation of Nucleotides:
The four types of nucleotides of DNA i.e. dAMP, dGMP, dCMP and dTMP are present in the nucleoplasm. They are activated by ATP in presence of an enzyme phosphorylase. This results in the formation of deoxyribonucleotide triphosphates i.e. dATP, dGTP, dCTP and dTTP. The process is known as Phosphorylation.
2. Point of Origin or Initiation point:
It begins at specific point 'O' -origin and terminates at point 'T'. Origin is flanked by 'T' sites. The unit of DNA in which replication occurs, is called replicon. In prokaryotes, there is ooly one replicon however in eukaryotes, there are several replicons in tandem. At the point 'O', enzyme endonuclease nicks one of the strands of DNA, temporarily. The nick occurs in the sugar-phosphate backbone or the phosphodiester bond.
3. Unwinding of DNA molecule:
Now enzyme DNA helicase operates by breaking weak hydrogen bonds in the vicinity of 'O'. The strands of DNA separate and unwind. This unwinding is bidirectional and continues as 'Y' shaped replication fork. Each separated strand acts as template.
The two separated strands are prevented from recoiling (rejoining) by SSBP (Single strand binding proteins).
SSB proteins remain attached to both the separated strands as to facilate synthesis of new polynucleotide strands.
4. Replicating fork:
The point formed due to unwinding and separation of two strands appear like a Y-shaped fork, called replicating/ replication fork.
The unwinding of strands imposes strain which is relieved by super-helix relaxing enzyme.
5. Synthesis of new strands:
Each separated strand acts as mould or template for the synthesis of new complementary strand.
It begins with the help of a small RNA molecule, called RNA primer. RNA primer get associated with the 3' end of template strand and attracts complementary nucleotides from surrounding nucleoplasm.
These nucleotides molecules bind to the complementary nucleotides on the template strand by forming hydrogen bonds (i.e. A=T or T=A; G = C or C = G).
The newly bound nucleotides get interconnected by phosphodiester bonds, forming a polynucleotide strand.
The synthesis of new complementary strand is catalyzed by enzyme DNA polymerase. The new complementary strand is always formed in 5'- 3' direction.
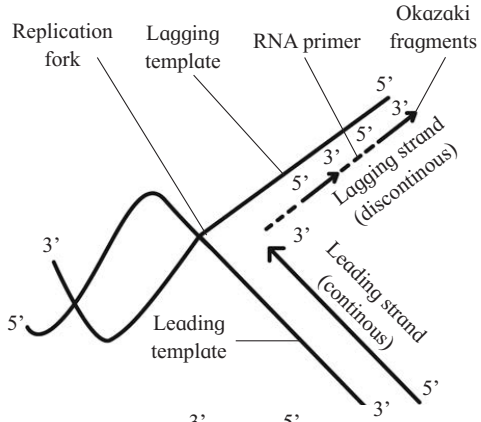
6. Leading and Lagging strand:
The template strand with free 3' end is called leading template and with free 5' end is called lagging template.
The process of replication always starts at C-3 end of template strand and proceeds towards C-5 end. As both the strands of the parental DNA are antiparallel, new strands are always formed in 5' → 3' direction.
One of the newly synthesized strand develops continously towards replicating fork is called leading strand.
Another new strand develop discontinuously away from the replicating fork is called lagging strand.
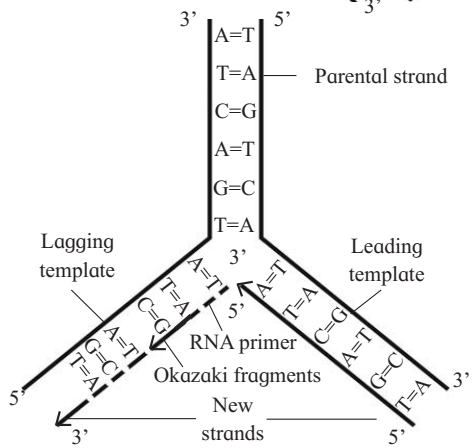
Maturation of Okazaki fragments :
DNA synthesis on lagging template takes place in the form of small fragments, called Okazaki fragments (named after scientist Okazaki).
Okazaki fragments are joined by enzyme DNA ligase. RNA primers are removed by DNA polymerase and replaced by DNA sequence with the help of DNA polymerase-I in prokaryotes and DNA polymerase-α in eukaryotes. Finally, DNA gyrase (topoisomerase) enzyme forms double helix to form daughter DNA molecules.
7. Formation of daughter DNA molecules:
At the end of the replication, two daughter DNA molecules are formed. In each daughter DNA, one strand is parental and the other one is totally newly synthesized. Thus, 50% is contributed by mother DNA. Hence, it is described as semiconservative replication.
Experimental confirmation : Semiconservative Replication :
In newly formed DNA molecule, one strand is old (i.e. conserved) and other strand is newly synthesized. Thus, it is called Semiconservative mode of replication.
It is experimentally proved by Matthew Meselson and Franklin Stahl (1958) by using equilibrium - density - gradient - centrifugation technique.
1. Meselson and Stahl in 1958 performed an experiment to prove semiconservative nature (mode) of replication.
2. They cultured bacteria E.coli in the medium containing 14N (light nitrogen) and obtained equilibrium density gradient band by using 6M CsCl. The position of this band is recorded.
3. E. coli cells were then tranferred to 15N medium (heavy isotopic nitrogen) and allowed to replicate for several generations. At equilibrium point density gradient band was obtained, by using 6M CsCl2. The position of this band is recorded.
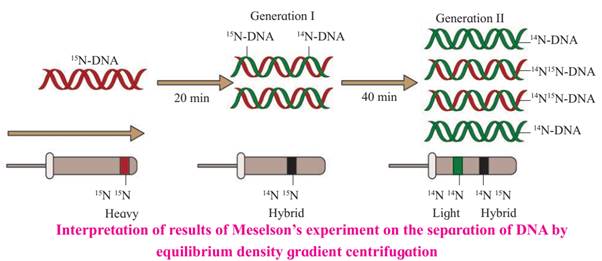
4. The heavy DNA (15N) molecule can be distinguished from normal DNA by centrifugation in a 6M Cesium chloride (CsCl2) density gradient. The density gradient value of 6M CsCl2 and 15N DNA is almost same. Therefore, at the equilibrium point 15N DNA will form a band. In this both the strands of DNA are labelled with 15N.
5. Such E. coli cells were they transferred to another medium containing 14N i.e. normal (light) nitrogen. After first generation, the density gradient band for 14N and 15N was obtained and its position was recorded. After second generation, two density gradient bands were obtained - one at 14N 15N position and other at 14N position.
6. The position of bands after two generations clearly proved that DNA replication is Semiconservative.
